Encoding and characters
Background
Detecting the encoding
Converting to UTF-8
Gremlins
graphu
Mojibake
Simplest characters
Combining characters
Markup and HTML entities
Quoting
Unmatched brackets
Cyrillic/Latin mixtures
Background
Datasets that will be shared with the world (like Darwin Core tables) should be in UTF-8 encoding. If you are not familiar with character encoding, here is a short backgrounder:
Computers only work with strings of 1s and 0s. "Encoding" is how a computer program knows what a particular string of 1s and 0s means, and how it should be displayed.
The character ö is what is called a two-byte character in UTF-8 encoding, because at the 1s and 0s level it contains two bytes (8 bits, each bit is a one or a zero). It looks like this: 11000011 10110110. Any program that understands UTF-8 encoding will see 11000011 10110110 and recognise that as ö.
In the Windows world, many programs do not understand UTF-8, and instead work with "Windows-1252" encoding, or CP1252. This is a single-byte encoding, which means that one byte is one character. In CP1252, the first byte 11000011 is à and the second byte 10110110 is ¶.
So, the two bytes 11000011 10110110 will be read as ö by a program that understands UTF-8, and as ö by a program that understands CP1252. The 1s and 0s have not changed, only the way they are interpreted.
The danger here is that a UTF-8 encoded file will be saved as CP1252, and then converted back to UTF-8. The converting program sees two characters in CP1252, à and ¶. It looks these up in its "table of conversions", and finds that à is what UTF-8 understands as the two-byte character 11000011 10000011, and ¶ is 11000010 10110110. This reproduces the incorrect ö in UTF-8 encoding and is a serious mistake.
It can get worse. Suppose the UTF-8 ö (now 11000011 10000011 11000010 10110110) is fed to a program that only understands CP1252. The program reads the four bytes one byte at a time, and sees à (11000011), ƒ (10000011),  (11000010), ¶ (10110110). The original string "E. Stöckhert" would now be "E. Stöckhert".
To avoid this confusion (called mojibake), always work with UTF-8: open files in a program that recognises UTF-8, and save the file in UTF-8 encoding.
Detecting the encoding
On the command line there are several tools available to test whether a text file is UTF-8 or not. The two I use regularly are file and uchardet.

"table1" does not have character encoding problems, because ASCII is included in UTF-8 . But the CRLF line endings must be changed to LF before any further data checking is done (see structure pages on "Carriage returns").
"table2" is ready for data checking.
"table3" should be converted to UTF-8 (see below), and then have its line endings changed before any further data checking is done.
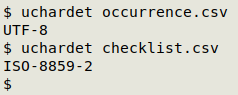
Like file, uchardet doesn't always get the encoding exactly right, but it is usually reliable. As shown in the screenshot below, "occurrence.csv" is OK, but "checklist.csv" is not in UTF-8 encoding and needs to be converted.
Converting to UTF-8
If you would prefer not to use the command line for conversion, consider using a text editor like Geany or Notepad++ to open the plain-text file and save it in UTF-8 with a new filename.
On the command line, the best all-purpose conversion tool is iconv. You should specify both the existing encoding (-f) and the target encoding, UTF-8 (-t utf-8). To convert a file from another encoding to UTF-8:
iconv -f [another encoding] -t utf-8 < [file] > [newfile]
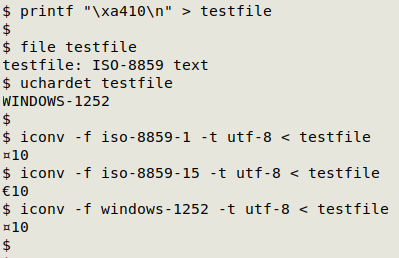
For a list of all the available "from" encodings, enter iconv -l. Unfortunately it can sometimes be hard to determine what the "from" encoding of your file might be. For example, the euro sign € has no encoding in ISO 8859-1 (an old encoding, but still used on some Windows computers). In ISO 8859-15, € is encoded as one byte with the hexadecimal value a4. file might say the encoding is ISO-8859, but which ISO-8859? And uchardet, like file, is sometimes wrong about the encoding. Watch what happens when I try to recover € with iconv:
Another iconv issue is that it stops converting when it finds an "illegal input sequence at position [some number]". It might be that the data table's encoding is corrupted, but a more likely explanation is that the "from" encoding being used is incorrect.
If you get the "illegal input sequence" message, you can try other likely "from" encodings. A more general solution is to find the "illegal input sequence" in the last line of the partly-converted table being built by iconv, with tail -1 [new file]. The (missing) character after the last one on the line must be the "illegal" one. It can be replaced with another character once its byte sequence is found in the original table. For details of this method see this BASHing data post.
If file or uchardet reports that the encoding is ASCII, it is not necessary to convert the data table to UTF-8. The entire ASCII character set is a subset of Unicode, on which UTF-8 is based. See also this BASHing data post for a truly strange encoding I found while checking a biodiversity data table.
Gremlins
In a Darwin Core table, all the characters should be either letters, numbers, ideographs, punctuation marks, spaces or newlines, and the only acceptable spaces are (horizontal) tabs and simple whitespaces.
"Gremlin" characters are different. They are invisible and can cause processing errors, and they should be deleted from the data table or converted to permitted characters. Three kinds of gremlins are detected with the gremlins script.
The first kind of gremlin is a control character. These are used by computers to manage text, and they can be dangerous in data processing. Many control characters get into data tables through incorrect encoding conversions and appear in mojibake. In other words, the person entering the data did not add the control character — it was added by a spreadsheet, database or other program which converted the data to another encoding.
A second kind of gremlin is the invisible space called "zero-width space", which I regularly find in Darwin Core tables.
A third kind of gremlin is a formatting character such as a no-break space (NBSP) or soft hyphen (SHY). These two characters are used by word-processing programs and Web browsers to manage how words are split over two lines. They are unnecessary in plain-text data tables and can easily cause data processing errors. NBSPs are particularly common in scientific names, for some reason.
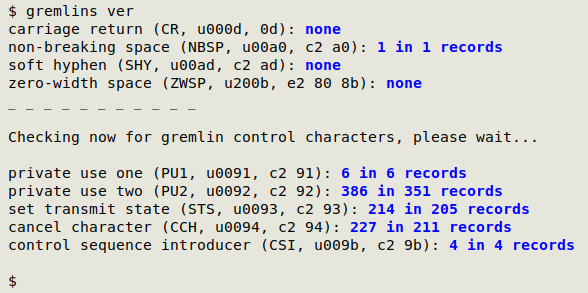
The screenshot below shows gremlins checking the file "ver". For each gremlin the script gives name, standard abbreviation, Unicode code ("uNNNN") and hexadecimal value. gremlins also detects carriage returns, which are covered separately on the structure page.
While the gremlins script tells you how many gremlins of each kind you have in a data table, it does not tell you where they are. For this purpose there are two command-line methods I can recommend. The first is the script gremfinderu (see scripts page). This not only shows you where the gremlins occur, but it also builds a text file on your desktop listing all the occurrences by line number in the data table. gremfinderu takes two arguments: filename and the Unicode code for each character ("uNNNN"), as shown below. You can get the "uNNNN" for each gremlin character from the gremlins script output (see screenshot above):

The second gremlin-finding method is noted below under mojibake (with charfind).
Gremlins are most easily deleted or replaced in a good text editor. For example, suppose you find with gremfinderu that the spaces in the scientificName entry "Aus bus Ivanov, 2014" are NBSPs, not plain spaces. Just copy the whole data item and paste it into the "search" box of a replacement dialog in the text editor, then enter the same data item with plain spaces in the "replace with" box and do "replace all". After doing your gremlin replacements and saving this new version of the data table, run gremlins again to make sure they are all gone.
There are also command-line methods for removing or replacing gremlins. Email me for suggestions on particular cases you find in your checking.
Occasionally a data table might have an invisible character that isn't detected by gremlins. The only ones I've seen in Darwin Core tables are the UTF-8 BOM and the left-to-right mark. These are detected by the graphu script but are harmless; they do not have to be deleted.
graphu
The graphu script tallies all the visible characters (and a few invisibles) in a file and also gives their Unicode code ("uNNNN").
Here graphu works on the file "char2"; the invisible u00ad character is a soft hyphen:
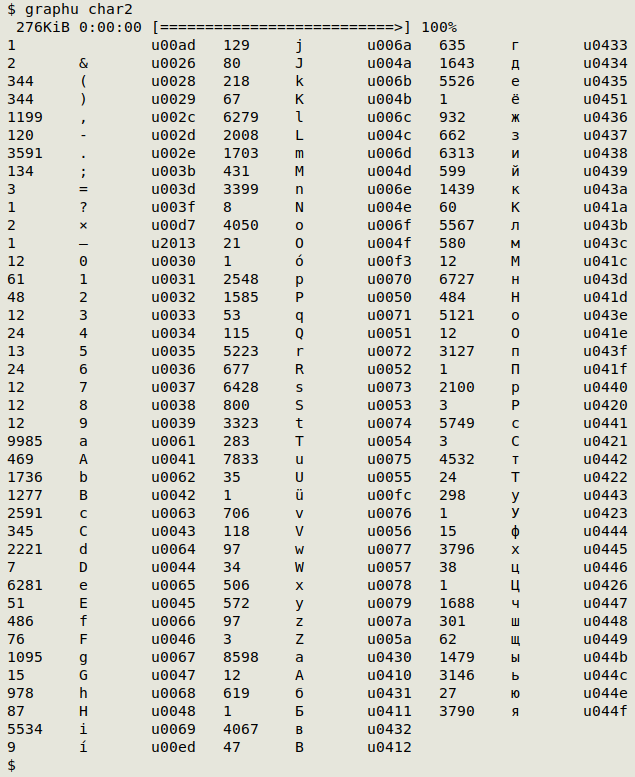
graphu can return thousands of characters if the dataset contains ideographs, as in some Chinese biodiversity datasets. A version of graphu called graphuNI is on the scripts page that omits ideographs from the character tally.
Mojibake
Conversion errors
Errors can occur when text data is moved between encodings. When the conversion is a failure, there are two possible results. One is that in the new encoding, the computer simply does not know how to interpret the 0s and 1s. You see � or "?" instead of the character you expect.
The second possible result is that the conversion goes ahead, but it goes ahead incorrectly. You see mojibake, like ö instead of ö. Sometimes there have been multiple "mojibakings", for example giving "d’Italia" when the original was "d’Italia" (see here).
Both of these two results are bad for data and need to be fixed. Not only do they corrupt some data items, they also make duplicates and pseudo-duplicates harder to find, where one data item is OK and the "same" data item contains an encoding conversion failure.
Uninterpretable results
Replacement characters (�) will appear in a graphu output, and you can also use the rcwords function to find them. rcwords takes the filename as its one argument.
rcwords() {
grep -o "[^[:blank:]]*["'\xef\xbf\xbd'"][^[:blank:]]*" "$1" | sort | uniqc | less -X
}
rcwords will sort and tally all the individual words containing � (if there are any). You can then use a text editor (or the command line) to replace a word with the correct one (see also below). Note the less -X in the function, which pages the output in your terminal but allows the output to persist there after less is closed with q.
The function qwords
qwords() {
grep -o "[^[:blank:]]*?[^[:blank:]]*" "$1" | grep -v "http" | sort | uniqc | less -X
}
does a similar job but tallies every word containing "?" except Web addresses (URLs). In this case you need to look carefully at the qwords output for likely encoding failures, such as "M?ller" where you would expect "Müller".
Mojibake
Unfortunately, there is no magic script or function that will find all the mojibake in a dataset. Instead, it is best to tally all the characters with graphu and look for characters not likely to be in the data table. There are certain characters that regularly appear in mojibake that has been caused by failed conversion from a Windows encoding to UTF-8. A very handy table showing these common failures is here.
Mojibake can also be spotted when you look through a tally of a field, and with experience you can recognise characters in a tally or graphu result that just "look wrong". When you spot such a character, you can locate it with either the charfind or charfindID functions:
charfind() {
echo "No. of records | Field name | Data item"; awk -F"\t" -v char="$(printf "\\$2")" 'NR==1 {for (i=1;i<=NF;i++) a[i]=$i} $0 ~ char {gsub(char,"\33[103m"char"\33[0m",$0); for (j=1;j<=NF;j++) if ($j ~ char) print a[j] FS $j}' "$1" | sort | uniqc | sed 's/\t/ | /g'
}
charfindID() {
echo "ID | Field name | Data item"; awk -F"\t" -v char="$(printf "\\$2")" -v idfld="$3" 'NR==1 {for (i=1;i<=NF;i++) a[i]=$i} $0 ~ char {gsub(char,"\33[103m"char"\33[0m",$0); for (j=1;j<=NF;j++) if ($j ~ char) print $idfld FS a[j] FS $j}' "$1" | sort -t $'\t' -k2 | sed 's/\t/ | /g'
}
charfind searches a file for a character by the character's Unicode code. It returns the data item containing the character (with the character highlighted in yellow), the field in which the data item appears, and the number of records with that data item.
charfindID lists the occurrences of the character and adds an identifier for the record from a unique ID field in the table.
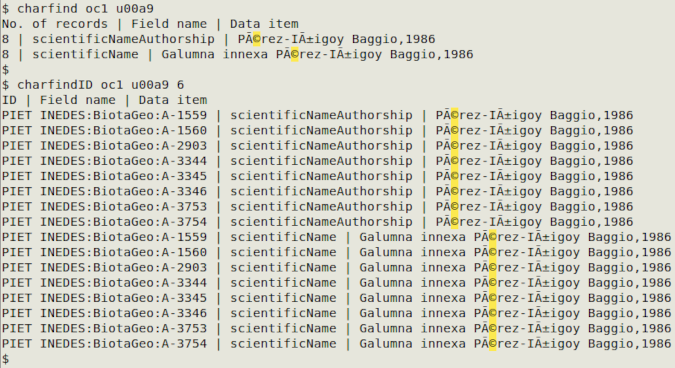
In the screenshot below, the two commands are looking for the © sign (u00a9) in the Darwin Core table "oc1". Field 6 in "oc1" contains occurrenceID, which is printed in the charfindID output as the first field.
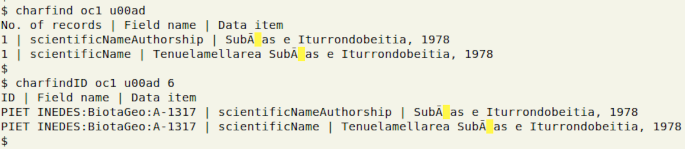
The two charfind functions can be used to pinpoint replacement characters (�), which have the Unicode code ufffd. Both functions can also find invisible characters, but these are only visualised as highlighted spaces. In the example below, I am looking for the soft hyphen gremlin (u00ad) in "oc1":
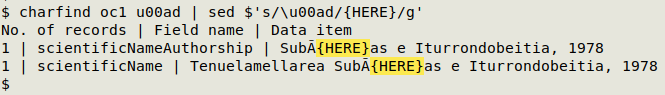
It's possible to replace the invisibles as Unicode codes with sed, but you need to use "quoted string expansion" by preceding the sed replacement code with "$":
Simplest characters
Darwin Core data compilers should use just one version of a character, namely the simplest one, especially if the data are not initially compiled in UTF-8. Using only the simplest form of a character not only avoids pseudo-duplicating data items but also makes encoding conversion failures less likely.
Examples of simplest versions:
- apostrophe ('), not left single quote (‘), right single quote (’) or grave accent (`)
- quotation mark ("), not left double quote (“) or right double quote (”)
- hyphen/minus (-), not en dash (–) or em dash (—)
- degree sign (°), not ring above (˚) or masculine ordinal indicator (º)
- quotation mark for minutes in DMS coordinates, not two apostrophes:
42°14'35"S, not 42°14'35' 'S - Unless you really need to have æ, use ae
You can see if there is more than one version of a character in a data table by running graphu and scanning the output. All instances of unwanted versions of a character can be replaced with a simpler version using a text editor, or on the command line with sed.
Combining characters
Combinations are a special case of "use the simplest version". Some characters can be UTF-8 encoded in more than one way. For example, there are two different ways to represent a Latin a with an umlaut (or diaeresis). You can code the single character ä, or you can code a plain a plus a diaeresis ¨. The isolated diaeresis character is a combining character.
In a Darwin Core table, always use a single UTF-8 character, not a plain character plus a combining character. The number of lines with combining characters (if any) can be found with the combocheck function, which applies a regular expression class with PCRE-enabled grep:
combocheck()
{
grep -cP "\p{M}" "$1"
}
If combining characters are detected with combocheck, the words containing them can be located by line number with:
awk -b '{for (i=1;i<=NF;i++) {if ($i ~ /\xcc[\x80-\xbf]/ || $i ~ /\xcd[\x80-\xaf]/) print NR,$i}}' [filename]
or the unique words tallied with:
awk -b '{for (i=1;i<=NF;i++) {if ($i ~ /\xcc[\x80-\xbf]/ || $i ~ /\xcd[\x80-\xaf]/) print $i}}' [filename] | sort | uniqc
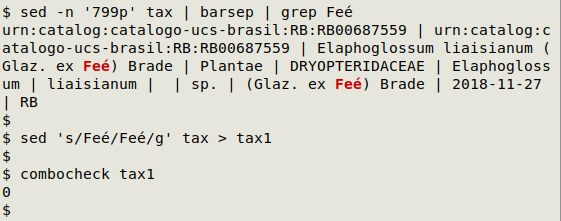
In the screenshot below, combocheck has found 1 line with combining characters in "tax", and the AWK command has found the line number (799) with the two words containing a combining character:

In "tax" the scientific author name Feé has been spelled with a plain e (u0065) and a combining acute accent (u0301). These two characters can be replaced with the single character é in a text editor, or on the command line using sed:
Markup and HTML entities
Darwin Core tables do not need and should not have any HTML markup, like "<i>" and "<b>", or HTML versions of characters, like "&" instead of "&". Markup often appears in reference citations, as in
Gàsser, Z. 1994. Un nou Clypeasteroid de l'Eocè català. <i>Batalleria</i> 4 (1990-1993), 13-16.
If only a few "<" and ">" are listed in the graphu output, you could check to see where these characters are and what they are doing (they could also represent "less than" and "greater than"). Use a grep search for line number:
grep -nE "<|>" [filename]
If there are many "<" and ">", try this command:
grep -nE "<.{,10}>" [filename]
This command searches for lines containing markup with at most 10 characters inside the "<" and ">". It will not only find markup, but also the strange character replacements sometimes produced by an R package, such as "M<fc>ller" instead of "Müller", where the replacement in angle brackets is the hexadecimal value of the Windows-1252 encoding for the character.
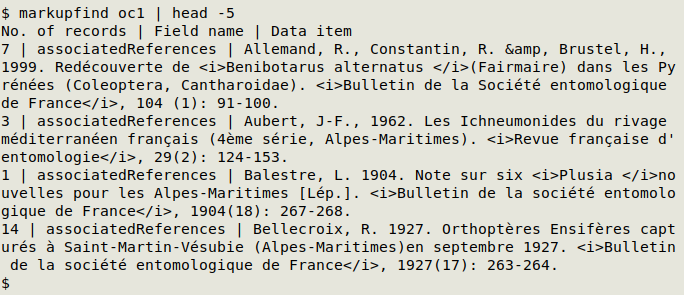
A more targeted way to search for markup is with the markupfind function:
markupfind() {
echo "No. of records | Field name | Data item"; awk -F"\t" 'NR==1 {for (i=1;i<=NF;i++) a[i]=$i; next} $0 ~ /<.{,10}>/ {for (j=1;j<=NF;j++) if ($j ~ /<.{,10}>/) print a[j] FS $j}' "$1" | sort | uniqc | barsep
}
markupfind tallies data items with markup and gives their fieldname as well. In the screenshot below, the first five results are shown for the Darwin Core table "oc1":
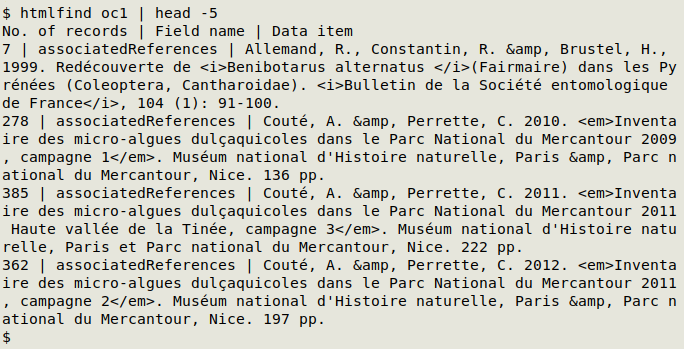
The corresponding function for HTML entities is htmlfind. In the screenshot below, htmlfind has found data items with "&":
htmlfind() {
echo "No. of records | Field name | Data item"; awk -F"\t" 'NR==1 {for (i=1;i<=NF;i++) a[i]=$i; next} $0 ~ /\t&[[:alnum:]]/ || $0 ~ / &[[:alnum:]]/ {for (j=1;j<=NF;j++) if ($j ~ /\t&[[:alnum:]]/ || $j ~ / &[[:alnum:]]/) print a[j] FS $j}' "$1" | sort | uniqc | barsep
}
Quoting
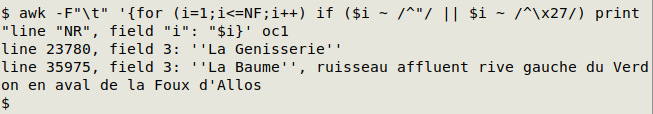
Darwin Core data tables (usually) do not need and should not have quotes around data items. Data items with unnecessary quotes often appear when a CSV file is converted to a data table, because data items with spaces or commas in CSVs are usually surrounded by quotes. It's easy to spot these items in a tally of an individual field. Alternatively, the following command will find all data items beginning with a quote (either ' or "), will give the line and field number, and will pipe the result to the less pager:
awk -F"\t" '{for (i=1;i<=NF;i++) if ($i ~ /^"/ || $i ~ /^\x27/) print "line "NR", field "i": "$i}' [filename] | less
Please note that some data items might have perfectly OK quotes, for example in these locality entries, where the quotes legitimately identify a placename:
5 km W of "The Springs"
"The Springs", Hamilton
In this check of "oc1", the command finds two data items where a placename is held between doubled apostrophes instead of between quotation marks:
Unmatched brackets
Check the graphu output for unmatched parentheses ( ) , square brackets [ ] and curly brackets { }. For example, there might be 2106 "(" but only 2103 ")". This mismatch in numbers can reveal two common problems. One is truncation, where a data item has been chopped off before the closing bracket:
ca 10 km E of Yekaterinburg (just SW of Ново-Свердловская ТЭЦ, ТГК № 9 power sta
The other common mismatch is a formatting mistake:
Erythronium sibiricum (Fisch. et C.A.Mey.] Krylov
Gàsser, Z. (1994 Un nou Clypeasteroid de l'Eocè català. Batalleria 4 (1990-1993), 13-16.
To locate data items containing unmatched brackets, use these three functions:
umparID() {
echo "ID | field name | data item"; awk -F"\t" -v idfld="$2" 'FNR==NR {if (NR==1) for (i=1;i<=NF;i++) a[i]=$i; else if (gsub(/\(/,"") != gsub(/\)/,"")) b[NR]; next} FNR in b {for (j=1;j<=NF;j++) if (split($j,c,"(") != split($j,d,")")) print $idfld FS a[j] FS $j}' "$1" "$1" | sort -t $'\t' -k2,2 -Vk1,1 | sed $'s/\t/ | /g;s/(/\033[103m(\033[0m/g;s/)/\033[103m)\033[0m/g'
}
umsquID() {
echo "ID | field name | data item"; awk -F"\t" -v idfld="$2" 'FNR==NR {if (NR==1) for (i=1;i<=NF;i++) a[i]=$i; else if (gsub(/\[/,"") != gsub(/\]/,"")) b[NR]; next} FNR in b {for (j=1;j<=NF;j++) if (split($j,c,"[") != split($j,d,"]")) print $idfld FS a[j] FS $j}' "$1" "$1" | sort -t $'\t' -k2,2 -Vk1,1 | sed $'s/\t/ | /g;s/[[]/\033[103m&\033[0m/g;s/[]]/\033[103m&\033[0m/g'
}
umcurID() {
echo "ID | field name | data item"; awk -F"\t" -v idfld="$2" 'FNR==NR {if (NR==1) for (i=1;i<=NF;i++) a[i]=$i; else if (gsub(/\{/,"") != gsub(/\}/,"")) b[NR]; next} FNR in b {for (j=1;j<=NF;j++) if (split($j,c,"{") != split($j,d,"}")) print $idfld FS a[j] FS $j}' "$1" "$1" | sort -t $'\t' -k2,2 -Vk1,1 | sed $'s/\t/ | /g;s/[{]/\033[103m&\033[0m/g;s/[}]/\033[103m&\033[0m/g'
}
Use umparID for parentheses, umsquID for square brackets and umcurID for curly brackets. The arguments for these functions are filename and number of a unique ID field, such as occurrenceID.
In the screenshot below, graphu shows unmatched parentheses in "taxon.txt". Next, umparID finds the data items (with parentheses highlighted in yellow), the name of the field with the data items and the taxonID (the ID field I chose) for that data item:
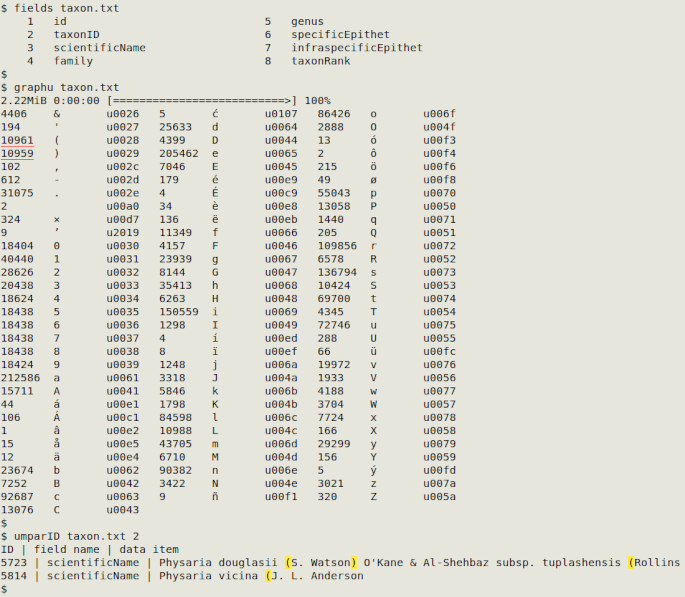
Cyrillic/Latin mixtures
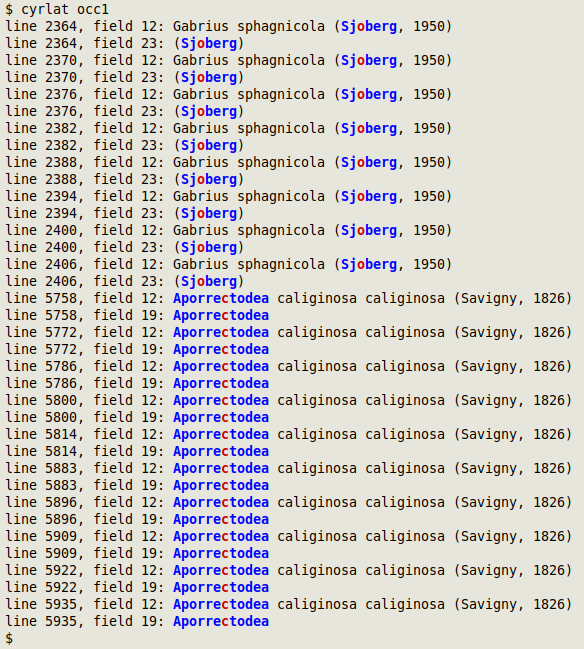
Some Darwin Core datasets have both Latin and Cyrillic words in various fields, and that isn't a problem. However, Cyrillic and Latin characters are sometimes mixed in individual words. This mixing can cause errors in data processing and also affect a search for duplicates or pseudo-duplicates. The cyrlat function finds any mixtures and colours Cyrillic in red and Latin in blue. The function takes filename as its one argument and prints line number, field number and the whole data item containing the mixed Cyrillic+Latin word.
cyrlat() {
latin=$(printf "[\\u0041-\\u005a\\u0061-\\u007a\\u00c0-\\u00ff\\u0160\\u0161\\u0178\\u017d\\u017e\\u0192]"); cyrillic=$(printf "[\\u0400-\\u04ff]"); awk -F"\t" -v lat="$latin" -v cyr="$cyrillic" '{for (i=1;i<=NF;i++) if ($i ~ cyr && $i ~ lat) print "line "NR", field "i": "$i}' "$1" | awk -v lat="$latin" -v cyr="$cyrillic" '{for (j=1;j<=NF;j++) {if ($j ~ cyr && $j ~ lat) {gsub(lat,"\033[1;34m&\033[0m",$j); gsub(cyr,"\033[1;31m&\033[0m",$j); print}}}' "$1"
}
In the screenshot below, cyrlat has checked the file "occ1" and found names in Latin characters but with Cyrillic "o" and "c":