Between records
Exact duplicates
Strict duplicates
UI duplicates
Relaxed duplicates
Exact duplicates
Exact duplicates have the same entries in all fields, including unique identifiers like occurrenceID. Exact duplicates probably come from bookkeeping errors when records are compiled, or when records are converted to Darwin Core format. They can be found with:
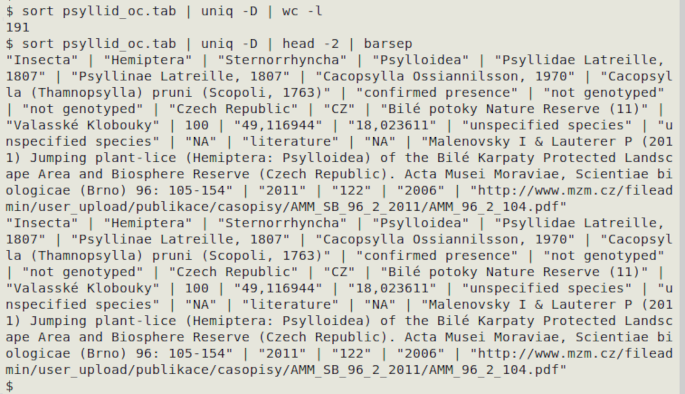
sort [filename] | uniq -D | wc -l
The output is piped to a line count to avoid flooding the terminal in case there are a lot of duplicates. The following example comes from an occurrence dataset and only the first two exact duplicates are shown:
Strict duplicates
Strict duplicates (my name for these!) have the same entries in all fields except those which give the record a unique ID, like the eventID field in an event.txt file. The following command finds strict duplicates and prints them as groups separated by a blank line:
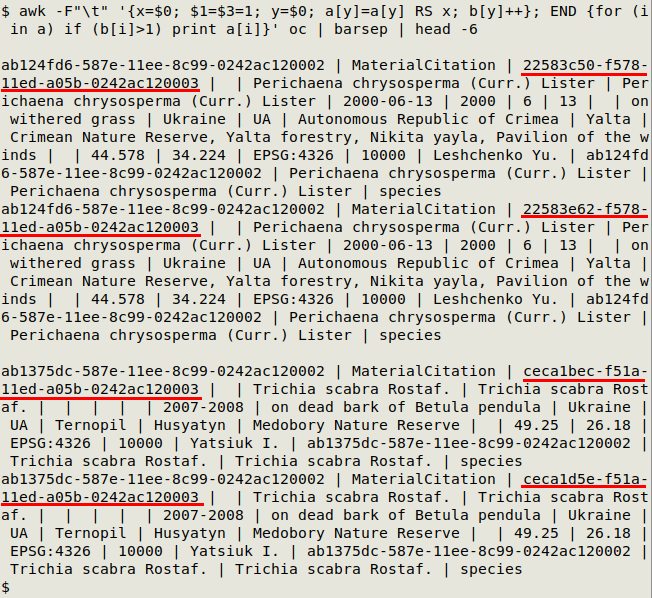
awk -F"\t" '{x=$0; $[UI field or fields]=1; y=$0; a[y]=a[y] RS x; b[y]++}; END {for (i in a) if (b[i]>1) print a[i]}' [filename]
In the screenshot below, I used both id and occurrenceID (underlined in red) as unique fields on "oc". Only the first two pairs of duplicates were printed. In this particular table id wasn't a unique ID field, however, so the strict duplicates differed only in occurrenceID.
UI duplicates
These are the opposite of strict duplicates: they are records that have the same UI entry (id, occurrenceID, eventID, measurementID, catalogNumber), but different entries in other fields. Because they are rare in biodiversity datasets, they are most easily found by looking first for duplicate UIs:
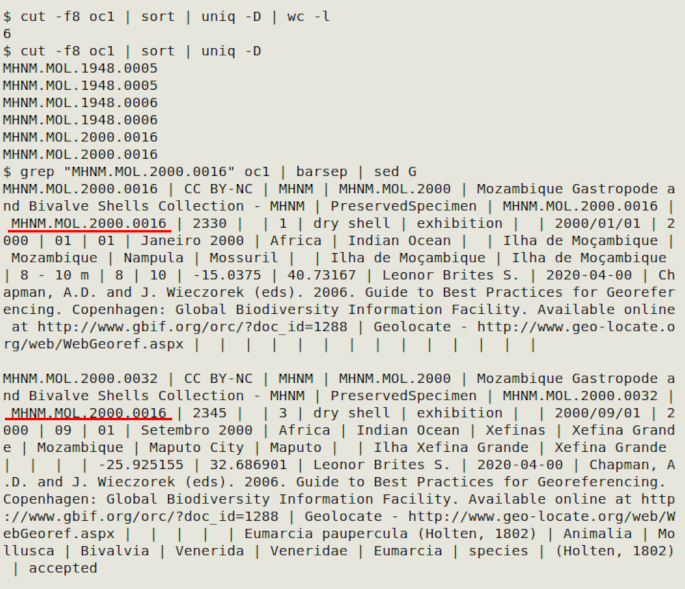
cut -f[UI field or fields] | sort | uniq -D | wc -l
In the screenshot below, the table "oc1" has three pairs of duplicate catalogNumber entries, but the other record details are different. I suspect in this case that catalogNumber was meant to be the same as id and occurrenceID, and there were data entry errors.
In the example I searched for records with one of the duplicated catalogNumber entries (underlined in red) using grep, and separated the "barsep"ed records with sed. A neater way to recover all records with a duplicated UI, separated into blank-line-separated groups, is with this command:
awk -F"\t" '{a[UI field number]=a[UI field number] RS $0; b[UI field number]++} END {for (i in a) if (b[i]>1) print a[i]}' [filename]
Relaxed duplicates
Relaxed duplicates (my name, again!) have the same entries in key fields, such as taxon, coordinates, date and collector/observer in an occurrence.txt table. These are most often not errors. For example, there might be several specimens of a particular insect species collected on the same day by the same person at the same location. Each specimen is on a different pin in the collection, and each has its own catalogNumber, so each has its own, unique record. The following command will find relaxed duplicates:
awk -F"\t" 'FNR==NR {a[fields]++; next} a[fields]>1' [filename] [filename]
"fields" means a comma-separated list of field numbers. For example, if the fields of interest in "oc1" are scientificName (field 16), decimalLatitude (68), decimalLongitude (69), eventDate (34) and recordedBy (32), then "fields" would be "$16,$68,$69,$34,$32":
awk -F"\t" '{a[$16,$68,$69,$34,$32]++; next} a[$16,$68,$69,$34,$32]>1' oc1 oc1
Note that this command will also pick up relaxed duplicates where entries in the selected fields are blank, because any blank is the same as any other blank. To only find records with non-blank entries, add conditions:
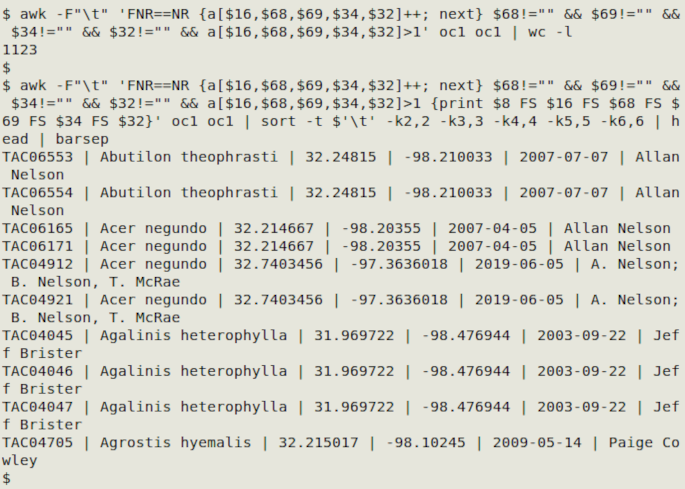
awk -F"\t" 'FNR==NR {a[$16,$68,$69,$34,$32]++; next} $68!="" && $69!="" && $34!="" && $32!="" && a[$16,$68,$69,$34,$32]>1' oc1 oc1
In the screenshot below, that last command found 1123 relaxed duplicate records. I modified the command as shown to print catalogNumber and each of the selected "relaxed" fields, then sorted the records by the selected fields and printed out the first 10 with barsep:
A much neater way to do this job involves a bit more coding, but it separates the relaxed duplicates into blank-line-separated groups and only requires one pass through the file by AWK:
awk -F"\t" '$68!="" && $69!="" && $34!="" && $32!="" {a[$16,$68,$69,$34,$32]=a[$16,$68,$69,$34,$32] RS $8 FS $16 FS $68 FS $69 FS $34 FS $32; b[$16,$68,$69,$34,$32]++} END {for (i in a) if (b[i]>1) print a[i]}' oc1
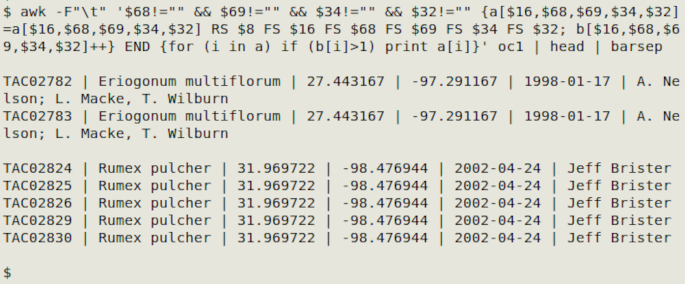
Below is a generic command for selected fields A, B, C. You can add other fields to be printed in the FS-separated series after "RS".
awk -F"\t" '$A!="" && $B!="" && $C!="" {a[$A,$B,$C]=a[$A,$B,$C] RS $A FS $B FS $C; b[$A,$B,$C]++} END {for (i in a) if (b[i]>1) print a[i]}' [filename]
Although the "blank-line-separated groups" output in the commands above is neat, you might like to sort it, for example by catalogNumber or scientificName. Email me for advice if you have a particular dataset you would like to filter and sort this way.
