
For a full list of BASHing data blog posts, see the index page. ![]()
Question marks that aren't really question marks
Some of the question marks ("?") in a dataset may not have been put there by the person or program entering the original data. They may have been entered later by a program that didn't understand the data's encoding. In other words, a question mark may be the "later" program saying I haven't a clue what this character is, so I'll replace it with a "?".
These replacement question marks are common in datasets with a complicated encoding history, like UTF-8 > windows-1252 > UTF-8. An example might be "Duméril" becoming "Dum?ril". But since a question mark is a valid character in most encodings, once inserted it's always a question mark. Unlike some replacement characters... it can't be converted back to an original character by changing the encoding. The original character is lost and can only be re-inserted after considering context, external sources or the advice of the data manager or custodian.
It's hard to detect these replacement question marks. The best way I know is to scan all the data items containing the "?" character and look for anomalous occurrences. In the Cookbook I describe a "qwords" function which lists all the words containing "?" (including isolated "?") and their frequencies. The list can be gigantic if there are fields in the dataset full of URLs with "?" in them, so URLs are excluded:
qwords() { grep -o "[^[:blank:]]*?[^[:blank:]]*" "$1" \
| grep -v "http" | sort | uniq -c; }
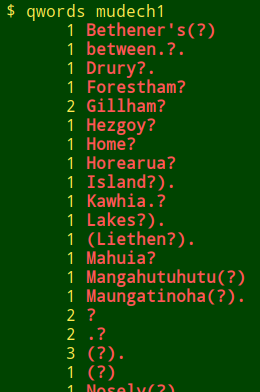
I also use a script called "qfinder" that tells me which fields contain non-URL "?", and on how many lines. The "qfinder" recipe is an improvement on one the Cookbook previously offered. It first generates the file "qlist-[filename]" in the working directory. This file lists all field contents containing "?" together with their field and line numbers, sorted first by field, then by line number. "qfinder" then asks me whether I'd like to see a uniquified version of "qlist-[filename]" (ignoring line numbers) using the less command:
#!/bin/bash
awk -F"\t" '{for (i=1;i<=NF;i++) {if ($i ~ /\?/ && $i !~ /http/) {print NR FS i FS $i}}}' "$1" | sort -t $'\t' -nk2 -nk1 > qlist-"$1"
declare -a label=($(head -n1 "$1" | tr '\t' '\n'))
qflds=$(cut -f2 qlist-"$1" | sort -n | uniq)
echo
echo -e "Table \"$1\" has \"?\" words in the following field(s):" \
for k in $(echo "$qflds"); do echo -e " field $k ($(echo ${label[$k-1]})) \
on $(awk -F"\t" -v fld="$k" '$2==fld' qlist-$1 | wc -l) lines"; done
echo
read -p "Show uniquified results with less? (y/n)" foo
echo
case $foo in
n) exit 0 ;;
y) less -fX <(cut -f2- qlist-"$1" | sort -n | uniq);;
esac
exit
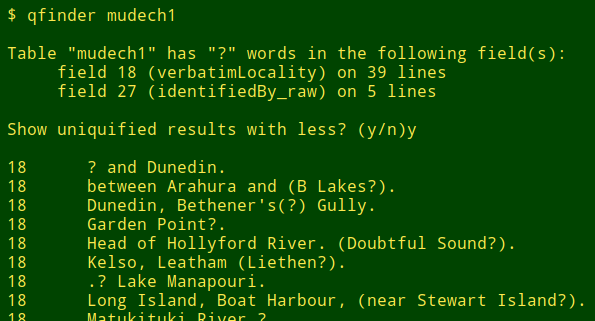
The less command in the "qfinder" script uses a couple of possibly unfamiliar options. The -f option allows less to deal with the "unreal" file from the process substitution <(cut -f2- qlist-"$1" | sort -n | uniq). The -X option keeps less from clearing the terminal screen; I can scroll back to whatever the terminal was showing before less wrote to stdout.
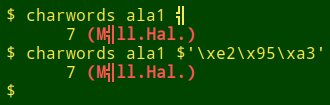
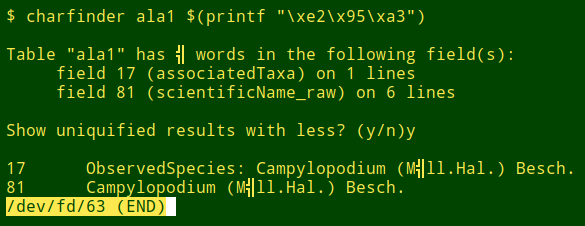
The same strategy could be applied to find any character in a data table, but it would mainly be useful for locating odd characters that really don't belong in the file. Below is a generalisation of "qwords" to "charwords". It's used here to find a very odd character in the file "ala1", then to find that odd character by its hex representation:
charwords() { grep -o "[^[:blank:]]*$2[^[:blank:]]*" "$1" | sort | uniq -c; }
To turn "qfinder" into "charfinder", the first AWK command needs to be tweaked (I've dropped here the URL exclusion):
Change
awk -F"\t" '{for (i=1;i<=NF;i++) {if ($i ~ /\?/ && $i !~ /http/) {print NR FS i FS $i}}}' "$1" \
| sort -t $'\t' -nk2 -nk1 > qlist-"$1"
to
awk -F"\t" -v char="$2" '{for (i=1;i<=NF;i++) {if ($i ~ char) {print NR FS i FS $i}}}' "$1" \
| sort -t $'\t' -nk2 -nk1 > qlist-"$1"
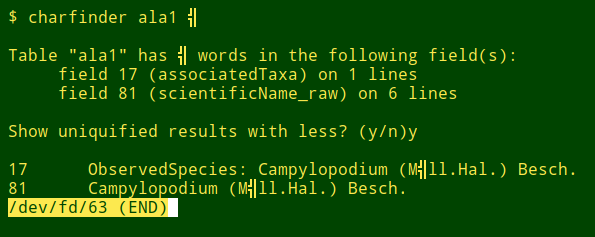
You could also run "charfinder" with the hex value of an odd character:
Last update: 2020-01-13
The blog posts on this website are licensed under a
Creative Commons Attribution-NonCommercial 4.0 International License